Understanding and Controlling Decisions
Monitoring the dynamics of the decision making process is the first step towards understanding how developmental and behavioral decisions are made. Measuring the dynamics of complex molecular and neural networks that control these decisions is challenging.The first direction of our research program has been to develop computational and experimental tools to rapidly identify key nodes within gene regulatory and neural networks whose dynamics control and best reflect the process of decision. The goal is to read the “mind” of the cell or animal as it makes a decision.
The second direction of the program is to monitor the decisions in real time to understand how individual multi-potent cells of the embryo receive and process signals to make developmental decisions, how signal reception and processing is affected by the geometry of the embryo, and how these decisions lead to the patterning of the embryo. These questions are focussed on addressing an open and important question in developmental biology: how the size of tissues and organs and the timing of their development are determined.
The third direction is to exploit our understanding to control the decisions that cells and animals make as well as to re-engineer the underlying networks to produce novel phenotypes. This this end, we want to develop and employ novel bioengineering, microscopy and optogenetic tools. We further exploit the cross species comparisons and analysis to achieve our goal.
Areas of Investigation
Discovering Key Nodes and Reading Minds of Gene Regulatory Networks
Modern high throughput approaches allow us to measure the levels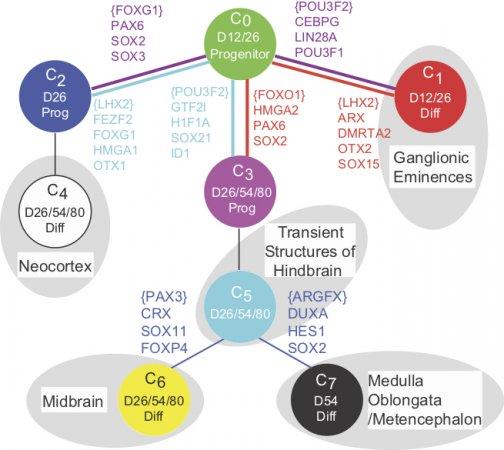 of thousands of genes in single cells of the embryo, or the activity patterns from thousands of neurons in freely moving animals. Such data provides novel challenges to conventional statistics because of low data density. Thus, identifying the small number key nodes whose dynamics reflect and control the process of decision making from these data is major challenge. An upcoming article from our lab explains and overcomes some of these challenges.
of thousands of genes in single cells of the embryo, or the activity patterns from thousands of neurons in freely moving animals. Such data provides novel challenges to conventional statistics because of low data density. Thus, identifying the small number key nodes whose dynamics reflect and control the process of decision making from these data is major challenge. An upcoming article from our lab explains and overcomes some of these challenges.Our approach has been (see our manuscript led by Furchtgott and Melton) to learn patterns of gene expression from known lineage decisions and exploit this pattern to analyze single cell gene expression data. Using a probabilistic approach, we thus identify key genes whose dynamics reflect the dynamics of decision making (watch multi-potent cells making decisions). This approach in turn allowed us to infer the sequence of lineage decisions giving rise to both excitatory and inhibitory neurons during early human brain development as reported in two manuscripts led by Yao and Close.
While our recent approaches allow us to detect important patterns in large data sets, a fundamental challenge that remains is to gain an understanding of the underlying networks from these large data sets. We want to develop causal predictive models of the underlying network from these data.
Signaling and Fate Decisions during early Human and Mouse Development.
The ability to watch cells make decisions has allowed us to as fundamental questions about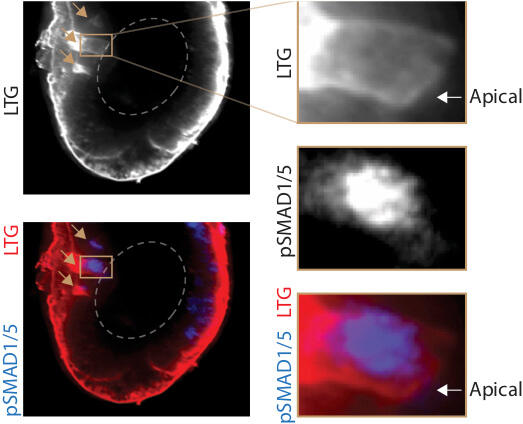 how cells receive signals, when and how they commit to a decision, how this decision is co-ordinated with the cell cycle and the fate choices of surrounding cells and how the geometry of the embryo affects patterning.
how cells receive signals, when and how they commit to a decision, how this decision is co-ordinated with the cell cycle and the fate choices of surrounding cells and how the geometry of the embryo affects patterning.We want to understand human embryonic stem cells and multipotent cells in the mouse embryo receive integrate signals. We discovered that all the BMP and TGF beta receptors in the embryo are basolaterally localized, and the LTA amino acid motif in their sequence governs this localization. In this study led by Zhechun Zhang and Steven Zwick, we also found that mislocalization of the receptors leads to mispatterning. The localization of the receptors in epithelial sheets of cells naturally leads to the coupling of the embryo geometry to patterning. This study leads to the question of how epithelial mesenchymal transition, signaling ligand diffusion and cell movement affect patterning by changing the geometry of the embryo.
The fate decision of a cell depends not only on what signals the multi-potent cells senses, but also when they sense it. The same BMP and TGF beta signals can cause pluripotent cells of the epiblast to adopt a mesendodermal, ectodermal or neural fate depending on when the cells sense the signal. Our ability to read the mind of the cell as it makes developmental decisions along the developmental trajectory, we can prospectively predict what fate each cell will adopt in response to signal. In this recent work led by Jim Valcourt, we exploit this predictive ability using RNA-seq and ATAC-seq to identify and validate candidate transcription factors that can modulate mesendoderm competence. These factors exert their effects by controlling the cell’s progression along the developmental trajectory, by tuning its competence to form mesendoderm at any given point along that trajectory, or by altering both of these aspects. In the classical picture of a Waddington landscape, these effects correspond to changing the cell’s location on the landscape and altering the location of the barrier between fates, respectively.
The ability of the underlying gene regulatory network to modulate these two aspects of the developmental landscape could allow separate control of the dynamics of differentiation and tissue size proportions. We are now exploiting microfluidic devices as in our prior work , human stem cell lines and microscopy to further understand how patterning and tissue proportions are established.
Comparative Analysis
Are there novel progenitor cell types associated with human and in particular cortical development?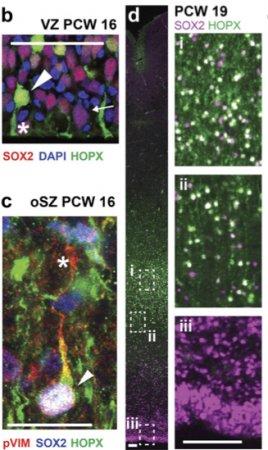 If so, how are these cell types generated and what do they give rise to? Can we use comparative genomics to rewire the underlying networks generate novel phenotypes?
If so, how are these cell types generated and what do they give rise to? Can we use comparative genomics to rewire the underlying networks generate novel phenotypes?To determine the molecular identities of progenitor cells in developing human fetal tissue, we developed single cell sequencing methods to extract intact RNA from fixed antibody stained cells from tissue (see Thomsen et al.). This was previously not possible because traditional methods of fixing, permeabilizing and antibody staining cells led to extensive RNA degradation.Through these studies we identified molecular markers for outer radial glial (oRG) progenitors that are enriched in the developing human cortex. We are now working to determine how these cell types arise and which neuronal and glial cell types they give rise to. Following previous work where we could exploit comparative analysis across multiple species to determine how to engineer signaling pathways (see Mody et al.), we are working towards determining how the underlying molecular networks that govern oRG generation.
Discovering key nodes in Neural Networks
Finding the key nodes of a complex network that govern specific behaviors is like finding a needle in a haystack. The conventional approach has been to look for these nodes one by one, which is challenging because such a search requires as many measurements as there are nodes, and because specific genetic tools that perturb only one node are often not available. We asked if we could search for the key nodes much more efficient with a novel experimental approach based on Compressed Sensing.
finding a needle in a haystack. The conventional approach has been to look for these nodes one by one, which is challenging because such a search requires as many measurements as there are nodes, and because specific genetic tools that perturb only one node are often not available. We asked if we could search for the key nodes much more efficient with a novel experimental approach based on Compressed Sensing.In a first such work led by Jeff Lee and Abdullah Yonar, we could infer the identity key interneurons from among the hundred interneurons in C.elegans, that controlled the speed of locomotion of the animal using just twenty eight measurements. In previous work by Zengcai Guo, we developed microscopy methods to optically perturb and measure calcium activity in animals that were confined. We extended this approach to by building a new microscope that could perturb neural activity in freely moving animals while monitoring their neural activity for more than an hour. Using this microscope. we could validate our inferences.
Our goal is to understand the nervous system sufficiently well to be able to take control of the animal’s behaviors. In work led by Askin Kocabas, we have been able to make the animal think that there is a chemo-attractive gradient in the environment and track this gradient (watch a short YouTube video of C. elegans searching for food). Using a combination of computation, microscopy and optogenetics, we are now trying to understand the different behaviors of the nematode and identifying neurons underlying learning and memory.
Technique Development: Optics and Microfluidics.
We have build microfluidics (Hersen and McClean, Nachman, Zhang and Zwick),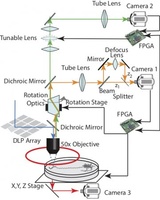 microscopy (Guo, Lee and Yonar, Kocabas) and image processing tools to be able to perform the experiments we want. Such tool building has become an integral part of our lab. We are currently collaborating with the Liu, Weitz and Mooney labs at Harvard to build new and improved tools.
microscopy (Guo, Lee and Yonar, Kocabas) and image processing tools to be able to perform the experiments we want. Such tool building has become an integral part of our lab. We are currently collaborating with the Liu, Weitz and Mooney labs at Harvard to build new and improved tools.
